
A/Prof Ulf Schmitz ~ Associate Professor- Bioinformatics; Associate Dean, Research
Biomedical Sciences and Molecular Biology
- About
-
- Teaching
- Interests
-
- Professional
-
- Mentoring Early Career Researchers
- Research
-
- RNA Biology
- Cancer Biology
- Bioinformatics
- Systems Biology
- Experience
-
- 2024 to present - Associate Dean Research, College of Public Health, Medical & Vet Sciences (JCU)
- 2023 to present - Section Head, Bioinformatics and Systems Biology, James Cook University
- 2021 to present - Associate Professor Bioinformatics, James Cook University
- 2018 to present - Associate Faculty, Centenary Institute (University of Sydney)
- 2017 to present - Hon Adj Professor, Chhattisgarh Swami Vivekanand Technical University (India)
- 2015 to present - Conjoint Senior Lecturer, Faculty of Medicine & Health (University of Sydney)
- 2018 to 2021 - Lab Head, Centenary Institute (University of Sydney)
- 2018 to 2021 - Senior Research Officer, Centenary Institute (University of Sydney)
- 2015 to 2017 - Research Officer, Centenary Institute (University of Sydney)
- 2003 to 2015 - Systems Engineer, University of Rostock (Germany)
- Research Disciplines
- Socio-Economic Objectives
Ulf is a computational biologist with training in bioinformatics and systems biology and more than 15 years of experience in analysing post-transcriptional gene regulation. His research interests focus on computational RNA biology and Systems Medicine.
Ulf completed his PhD at the University of Rostock (Germany). In 2015, he joined the Centenary Institute in Sydney as a post-doctoral Research Officer in the Gene and Stem Cell Therapy Program. He later established the Computational Biomedicine Lab at the Centenary. He also holds an appointment as Conjoint Senior Lecturer at the University of Sydney (Faculty of Medicine & Health).
Ulf and his team develop integrative workflows combining various computational disciplines with experimentation to address questions around non-coding RNAs, post-transcriptional gene regulation, and cancer biology. He has published 58 research articles, 11 book chapters, and 4 books on this topic.
Available HDR Student Projects
Single-cell data analysis of hepatocellular carcinoma patient samples
Portable long-read sequencing to diagnose Chronic Kidney Disease
Determining cellular gene isoform abundance from long-read RNA sequencing data
Mirtron synthesis and expression in leukemia
Mathematical Modelling of Alternative Splicing and Transcriptomic Complexity
Mathematical Modelling of Cell-Fate Determination
Computational Biomedicine Lab


In the Computational Biomedicine Lab at James Cook University and the Centenary Institute (University of Sydney) computational biologists work hand-in-hand with molecular biologists integrating in silico, in vitro and in vivo approaches in advanced interdisciplinary research. We develop integrative systems medicine workflows along with tools and databases to identify and study interactions between RNAs and other molecules, functional mechanisms of gene regulation, cell signalling pathways and gene regulatory networks.
Our goal is to achieve a deeper understanding of processes involved in disease emergence and progression, with a focus on cancer biology, and possible avenues for therapeutic interventions and optimised treatment schedules. Most projects involve the analysis of experimental high-throughput data (incl. (sc)RNA-seq, NanoString, DNA Methyl-Seq, ChIP-Seq, CLIP-seq, and more). Moreover, we use various mathematical formalisms to model the effects of environmental stimuli or pathogenic events on molecular interaction networks and signalling pathways. Using machine learning and computer simulation approaches, we predict disease progression and outcome as well as the effect of therapeutic agents on cellular survival and resistance mechanisms.
Current Research Projects
Portable long-read sequencing to diagnose Chronic Kidney Disease
With the help of new seed funding from the Tropical Australian Academic Health Centre, we trial a portable, rapid, and targeted genomic test to find genetic variants that predispose people towards Chronik Kidney Disease. For more information see recent media coverage:
Intron retention regulation in haematopoietic cells
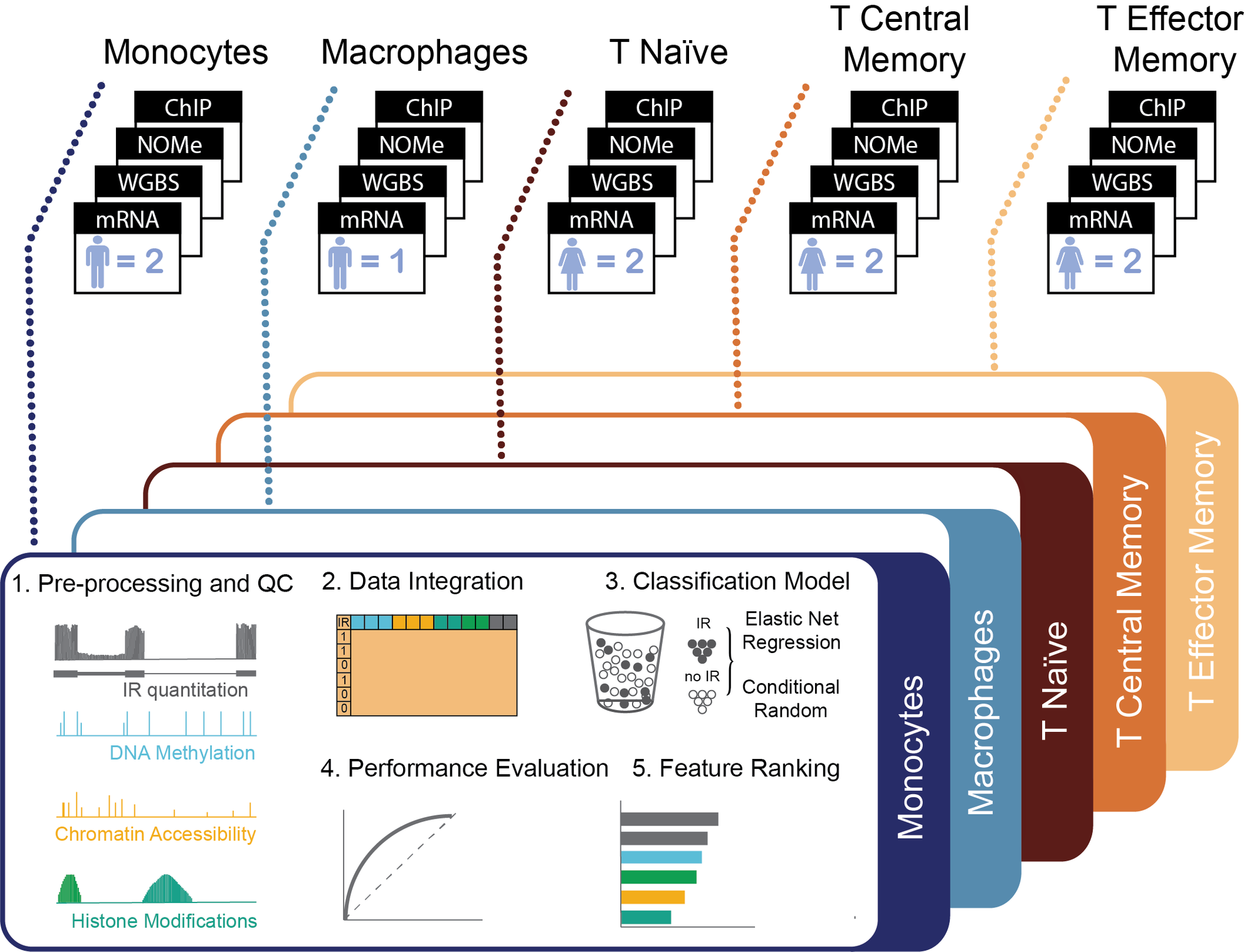
There is growing evidence that alternative splicing, including intron retention (IR), is regulated on at least two levels: locally, through a network of interacting trans-acting splicing regulators with cis-acting regulatory elements, and globally, through the chromatin structure. In this project, we explore a global regulation of IR in haematopoietic cells – we use statistical analysis to better understand IR regulation using information derived from the epigenetic factors that govern chromatin organisation, namely nucleosome assembly, DNA methylation, and histone modifications. The main challenge here is to integrate multiple layers of ‘-omics’ into a single computational model which produces biologically interpretable results. Find out more: Petrova et al, Nucleic Acids Research 2022
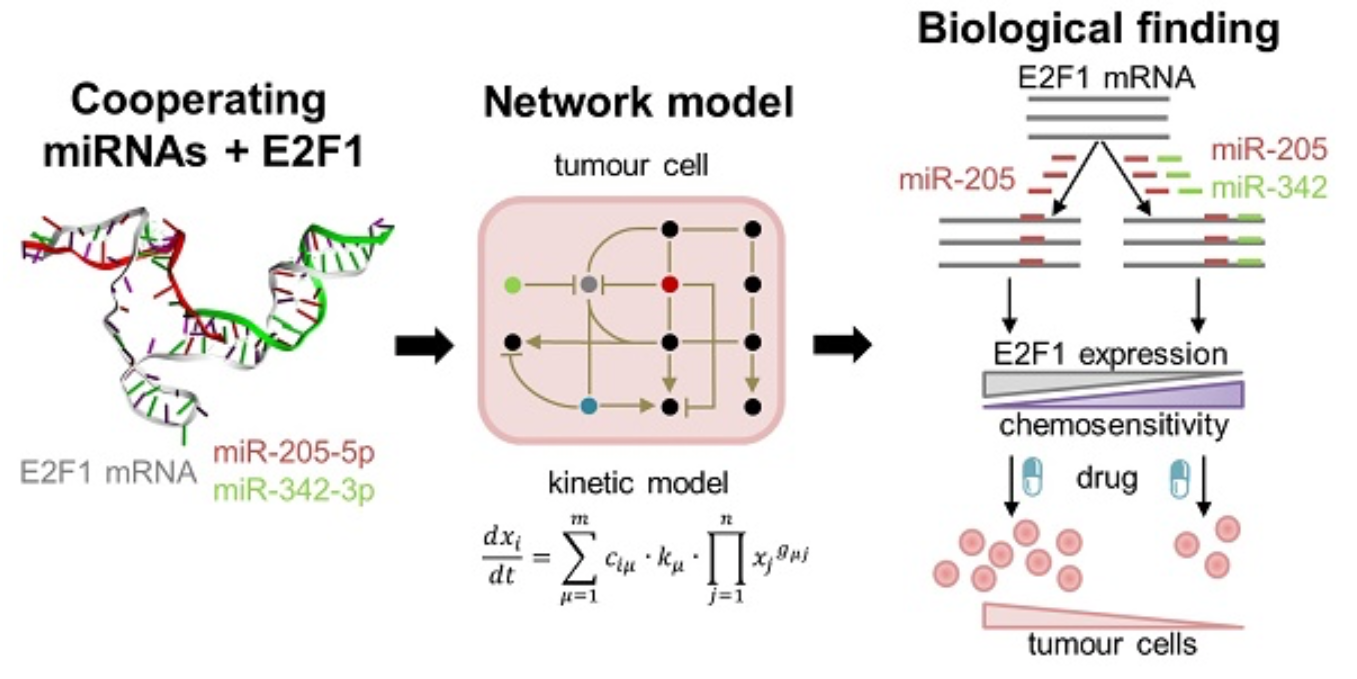
Cooperating microRNAs for cancer therapy
In this project, we use a systems medicine approach to find new avenues for overcoming chemotherapy resistance in aggressive tumour cells. Our results suggest that pairs of cooperating microRNAs could be used as potential RNA therapeutics to reduce E2F1-related chemoresistance. Find out more: Lai X, Gupta SK, Schmitz U et al, Theranostics 2018
Alternative splicing and the epigenome in Chronic Myeloid Leukaemia
In this project, we are investigating gene regulatory processes in leukaemia. The third biggest cause of cancer death in all Australians is blood cancers (leukaemia), which are diagnosed 35 times each day. Using a multi-omics approach, we examine alternative splicing and epigenetic changes in blood samples from chronic myeloid leukaemia (CML) patients before and after treatment with tyrosine kinase inhibitors. Find out more: Schmitz et al, Cancers 2020

Cross-talk between post-transcriptional gene regulation mechanisms
Combining computational predictions and in vitro assays, we elucidate the cross-talk between post-transcriptional gene regulation mechanisms. More specifically, we focus on interactions between microRNA molecules and intron-retaining mRNA transcripts and try to identify networks of interconnected gene regulation. Using a systems biology approach we will model competitive post-transcriptional gene regulation through iterative cycles of time-course experiments and model simulations.

MicroRNAs as Cancer Biomarkers
MicroRNAs are small molecules that play an important role in regulating gene expression. We have been exploring the potential of microRNAs as cancer biomarkers, as regulators of cancer genes and their role in immune cells. For more information about our work see recent media coverage:
- Honours
-
- Awards
-
- 2020 - Travel Award - The CASS Foundation
- 2020 - Centenary Recognition Award for Contribution to Centenary Life
- 2016 - Cure the Future Award of Scientific Excellence, Cure the Future Foundation
- 2016 - Joachim-Jungius Award for best PhD thesis at the University of Rostock, Germany
- Fellowships
-
- 2021 to 2025 - NHMRC Investigator (EL1)
- 2018 to 2020 - Cancer Institute NSW Early Career Fellowship
- 2018 to 2019 - Australia-India Strategic Research Fund (AISRF) Early and Mid-Career Fellowship, Australian Academy of Science
- 2017 to 2019 - NHMRC Peter Doherty Fellowship for Early Career Researchers
- Memberships
-
- 2023 - Editorial Board Member: Genome Biology
- 2021 - Topics board editor: Cancers
- 2019 - Editorial Board Member: Mathematical Biosciences and Engineering
- Other
-
- 2017 - Honorary Adjunct Professorship at Chhattisgarh Swami Vivekanand Technical University (CSVTU), Bhilai, India
- 2016 - Conjoint Senior Lecturer, Sydney Medical School, University of Sydney
- Publications
-
These are the most recent publications associated with this author. To see a detailed profile of all publications stored at JCU, visit ResearchOnline@JCU. Hover over Altmetrics badges to see social impact.
- Journal Articles
-
- Dorney R, Dhungel B, Rasko J, Hebbard L and Schmitz U (2023) Recent Advances in Cancer Fusion Transcript Detection. Briefings in Bioinformatics, 24 (1).
- Gillman R, Field M, Schmitz U, Karamatic R and Hebbard L (2023) Identifying cancer driver genes in individual tumours. Computational and Structural Biotechnology Journal, 21. pp. 5028-5038
- Oehler J, Wright H, Stark Z, Mallett A and Schmitz U (2023) The application of long?read sequencing in clinical settings. Human Genomics, 17.
- Sadeghi M, Karimi M, Karim A, Farshbaf N, Barzegar A and Schmitz U (2023) Network-Based and Machine-Learning Approaches Identify Diagnostic and Prognostic Models for EMT-Type Gastric Tumors. Genes, 14 (750).
- Thavaneswaran S, Kansara M, Lin F, Espinoza D, Grady J, Lee C, Ballinger M, Sebastian L, Corpuz T, Qiu M, Mundra P, Bailey C, Schmitz U, Simes J, Joshua A and Thomas D (in press) A signal-seeking Phase 2 study of olaparib and durvalumab in advanced solid cancers with homologous recombination repair gene alterations. British Journal of Cancer,
- Wong A, Wong J, Rasko J and Schmitz U (2023) SpliceWiz: Interactive analysis and visualization of alternative splicing in R. Briefings in Bioinformatics, 25 (1).
- Wu S and Schmitz U (2023) Single-cell and long-read sequencing to enhance modelling of splicing and cell-fate determination. Computational and Structural Biotechnology Journal, 21. pp. 2373-2380
- Karimi M, Karimi A, Abolmaali S, Mehdi S and Schmitz U (2022) Prospects and challenges of cancer systems medicine: from genes to disease networks. Briefings in Bioinformatics, 23 (1).
- Petrova V, Song R, DEEP Consortium , Nordström K, Walter J, Wong J, Armstrong N, Rasko J and Schmitz U (2022) Increased chromatin accessibility facilitates intron retention in specific cell differentiation states. Nucleic Acids Research, 50 (20). pp. 11563-11579
- Shah J, Milevskiy M, Petrova V, Au A, Wong J, Visvader J, Schmitz U and Rasko J (2022) Towards resolution of the intron retention paradox in breast cancer. Breast Cancer Research, 24.
- Book Chapters
-
- Schmitz U (2023) Overview of Computational and Experimental Methods to Identify Tissue-Specific MicroRNA Targets. In: MicroRNA Detection and Target Identification. Methods in Molecular Biology, 2630. Humana Press, New York, NY, USA, pp. 155-177
- Lai X, Schmitz U and Vera J (2022) The Role of MicroRNAs in Cancer Biology and Therapy from a Systems Biology Perspective. In: The Role of MicroRNAs in Cancer Biology and Therapy from a Systems Biology Perspective. Advances in Experimental Medicine and Biology, 1385. Springer, Cham, pp. 1-22
- More
-
ResearchOnline@JCU stores 50+ research outputs authored by A/Prof Ulf Schmitz from 2007 onwards.
- Current Funding
-
Current and recent Research Funding to JCU is shown by funding source and project.
Townsville Hospital and Health Service - SERTA Research Capacity Grant
Improving outcomes for patients with muscle wasting disease and liver cancer - SERTA Capacity Research Grant.
- Indicative Funding
- $140,000 over 2 years
- Summary
- The funding of a Research Assistant and the performance of work by the Research Assistant in connection with a program of work undertaken by the Cancer and Metabolism Group within the College of Public Health, Medicine and Veterinary Science aimed at improving outcomes for patients with muscle wasting disease and liver cancer.
- Investigators
- Pankaj Saxena, Lionel Hebbard, Jaishankar Raman, Rozemary Karamatic, Matan Ben David, Craig McFarlane, Ulf Schmitz, Matt Field and Miriam Wankell in collaboration with Shaurya Jhamb, Eun Jin Sun, Zaeem Ahmed, Rhys Gillman and Ryley Dorney (Townsville Hospital and Health Service, College of Public Health, Medical & Vet Sciences and College of Medicine & Dentistry)
- Keywords
- Sarcopenia; Hepatocellular Carcinoma; Liver Cancer
Townsville Hospital and Health Service - Study Education Research Trust Account (SERTA)
Establishing the North Queensland Liver Tumour Library: Improving HCC detection and treatment.
- Indicative Funding
- $50,000 over 2 years (administered by Townsville Hospital and Health Service)
- Summary
- The project will provide certain data on the usefulness of using liquid biopsy to diagnose HCC in Rural, regional and Remote patients, identify new liquid biopsy targets and test a new therapeutic approach to treat HCC.
- Investigators
- Rozemary Karamatic, Matan Ben David, Pankaj Saxena, Lionel Hebbard, Craig McFarlane, Ulf Schmitz, Matt Field and Miriam Wankell in collaboration with Shaurya Jhamb, Eun Jin Sun, Zaaem Ahmed, Rhys Gillman and Ryley Dorney (Townsville Hospital and Health Service, College of Public Health and Medical & Vet Sciences)
- Keywords
- Liquid biopsy; Hepatocellular Carcinoma; Liver Cancer
National Health & Medical Research Council - Investigator Grants
Investigating post-transcriptional gene regulation in cancer
- Indicative Funding
- $577,910 over 4 years
- Summary
- In this project we willl employ integrative systems medicine approaches to generate a detailed mechanistic overview of multi-level cancer gene regulation and provide novel avenues for the treatment of aggressive tumours. We will determine patterns of gene regulation in leukaemia and test whether these patterns are predictive for disease outcome. We will devise strategies for the therapeutic interference with gene regulation to effectively sensitise aggressive tumours to chemotherapy and thereby diminish tumour cell populations and prevent relapse.
- Investigators
- Ulf Schmitz (College of Public Health and Medical & Vet Sciences)
- Keywords
- Bioinformatics; Breast Cancer; Gene Regulation; Computational Biology; Leukaemia; RNA biology
Tour de Cure - PhD Scholarship
Precision Hepatocellular Carcinoma Treatment Using Synthetic Lethality
- Indicative Funding
- $10,000 over 1 year
- Summary
- Hepatocellular carcinoma (HCC) is a major cause of cancer-related mortality. Current therapies for HCC show little efficacy due to extensive heterogeneity in the disease and a lack of corresponding patient-tailored treatment options. The purpose of this project is to validate various bioinformatic approaches to identifying personalised cancer driver genes and methods of targeting them through synthetic lethality. An integrated pipeline will be developed for the simple identification of therapeutics from sequencing of tissue biopsies. Finally, patient-derived organoids matched with patient-specific sequencing information will be used to validate this pipeline, building an important foundation for the future of personalised treatment.
- Investigators
- Rhys Gillman, Lionel Hebbard, Matt Field and Ulf Schmitz (College of Public Health and Medical & Vet Sciences)
- Keywords
- Liver Cancer; Genetics; Synthetic Lethality; Bioinformatics; Precision Medicine
Townsville Hospital and Health Service - SERTA Research Capacity Grant
Improving outcomes for patients with muscle wasting disease and liver cancer
- Indicative Funding
- $135,439 over 2 years
- Summary
- This application is to provide support for an experienced research assistant to perform laboratory work with human tissues. The interdisciplinary project involves TUH surgeons, junior medical staff, JCU/AITHM researchers, medical and HDR students; and pursues two themes to develop better diagnostics and treatments for patients with (i) the muscle wasting disease sarcopenia, and (ii) hepatocellular carcinoma (HCC). Human ethics for the collection and culturing of human muscle cells and HCCs has been approved by THHS and JCU committees. The Research Program focus is to sequence DNA of the primary muscle cells and HCCs, and use state of the art bioinformatics to identify and characterise novel biomarkers and molecular targets that drive sarcopenia and HCC progression. This will support the development of patient-focussed therapeutic approaches and provide better health care outcomes for North Queenslanders.
- Investigators
- Pankaj Saxena, Jaishankar Raman, Rozemary Karamatic, Matan Ben David, Lionel Hebbard, Craig McFarlane, Ulf Schmitz, Matt Field and Miriam Wankell (Townsville Hospital and Health Service, College of Public Health and Medical & Vet Sciences)
- Keywords
- Sarcopenia; Hepatocellular Carcinoma; Liver Cancer
Tropical Australian Academic Health Centre Limited - Research Seed Grants
Using portable long-read sequencing to diagnose Chronic Kidney Disease in regional North QLD Using portable long-read sequencing to diagnose Chronic Kidney Disease in regional North Using portable long-read sequencing to diagnose Chronic Kidney Disease
- Indicative Funding
- $50,000 over 2 years
- Summary
- This project addresses the early diagnosis of genetic predispositions for Chronic Kidney Disease (CKD), which poses an increasing burden on the North Queensland population and unequally affects the Indigenous Australian peoples. We will develop a mobile diagnostic pipeline that allows a rapid and cost-efficient screening for genetic CKD predispositions and circulating biomarkers using a targeted, DNA/RNA long-read sequencing approach.
- Investigators
- Ulf Schmitz, Andrew Mallett, Matt Field, Paul Horwood, Helen Wright, Chirag Patel, Ira Cooke and Ben Lundie (College of Public Health, Medical & Vet Sciences, College of Medicine & Dentistry, College of Healthcare Sciences, Queensland Clinical Genetics Service and Pathology Queensland)
- Keywords
- long-read sequencing; Nanopore; Chronic Kidney Disease; Genetic Testing; targeted sequencing; transcriptomic complexity
Cancer Council NSW - Project Grants
Deciphering cancer gene regulation
- Indicative Funding
- $404,943 over 4 years
- Summary
- In this project, we investigate regulatory processes in cancer, whereby the control of protein generation from DNA genetic code is disrupted. This process is referred to as intron retention (IR), a phenomenon, which allows ?junk? DNA to enter the cell. IR plays a critical role in cancer development, yet the mechanism of its involvement remains unresolved. We hypothesise that IR can significantly interfere with other forms of gene regulation via `cross-talk?. This introduces a distortion of gene regulation, which is amenable to therapeutic manipulation. Our enhanced understanding of gene-regulatory cross-talk will facilitate improved IR-directed therapies.
- Investigators
- Ulf Schmitz and Charles Bailey (College of Public Health, Medical & Vet Sciences and Centenary Institute of Cancer Medicine & Cell Biology)
- Keywords
- Bioinformatics; Breast Cancer; Gene Regulation; Computational Biology; Leukaemia; RNA biology
- Supervision
-
Advisory Accreditation: I can be on your Advisory Panel as a Primary or Secondary Advisor.
These Higher Degree Research projects are either current or by students who have completed their studies within the past 5 years at JCU. Linked titles show theses available within ResearchOnline@JCU.
- Current
-
- Clinical applications of genetic testing, sequencing and information to promote kidney health (PhD , Secondary Advisor/AM)
- Investigating monocyte and NK derived MS genes by scSequencing, flow cytometry and cytotoxicity assays. (PhD , Secondary Advisor)
- Insights into the molecular bases of coral-specific traits (PhD , Secondary Advisor)
- Computational approaches to Detect Genetic Variants at Single Cell resolution (PhD , Secondary Advisor)
- Neural network based model for cell type-specific isoform expression pattern (Masters , Primary Advisor/AM/Adv)
- Targeting DNA Repair in HCC through Synthetic Lethality (PhD , Secondary Advisor)
- Studies in the immune response to lung disease: rural PNG and urban Australia (PhD , Secondary Advisor)
- Developing a new treatment regime for hepatocellular carcinoma (PhD , Secondary Advisor)
- Analysing the role of chimeric transcripts in human cancers (PhD , Primary Advisor/AM/Adv)
- Pharmacogenomic Profiling of Mental Health Patients in the Philippine National Center for Mental Health (PhD , Secondary Advisor)
- Collaboration
-
The map shows research collaborations by institution from the past 7 years.
Note: Map points are indicative of the countries or states that institutions are associated with.- 5+ collaborations
- 4 collaborations
- 3 collaborations
- 2 collaborations
- 1 collaboration
- Indicates the Tropics (Torrid Zone)
Connect with me
- Phone
- Location
-
- 142.302, The Science Place (Townsville campus)
- Advisory Accreditation
- Advisor Mentor
- Find me on…
-






My research areas
Similar to me
-
Dr Alexandra TrollopeMedicine
-
A/Prof Lionel HebbardBiomedical Sciences and Molecular Biology
-
Dr Craig McFarlaneBiomedical Sciences and Molecular Biology
-
Dr Margaret JordanBiomedical Sciences and Molecular Biology
-
Dr Kishani TownshendCairns Institute







